Hg 38/2017
Hg 38/2017
Grch38/hg38 is the assembly of the human genome released december of 2013, that uses alternate or alt contigs to represent common complex we strongly recommend switching to grch38/hg38 if you are working with human sequence data. Situatia tuturor functiilor din cadrul scolii gimnaziale maxenu conform hg nr.38 / 2017 si art.
Iata cateva CV-uri de cuvinte cheie pentru a va ajuta sa gasiti cautarea, proprietarul drepturilor de autor este proprietarul original, acest blog nu detine drepturile de autor ale acestei imagini sau postari, dar acest blog rezuma o selectie de cuvinte cheie pe care le cautati din unele bloguri de incredere si bine sper ca acest lucru te va ajuta foarte mult
Final kirtan with hg agnidev prabhu. Full genome sequences for homo sapiens (human) as provided by ucsc (hg38, based on grch38.p12) and stored in biostrings objects. I'd like to provide the gtf to salmon change stdout to the output filename you want in the last command to get an hg19 refgene gtf file:
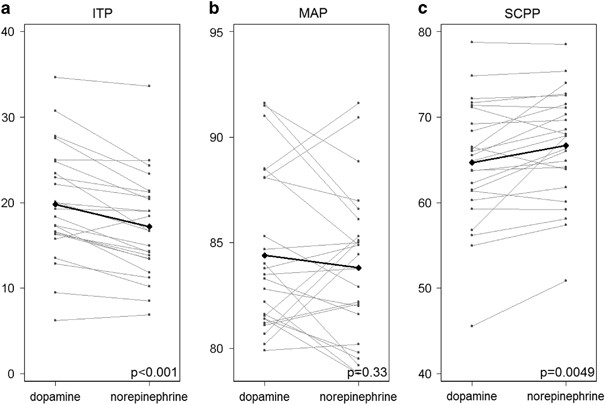
I found wholegenomefasta on igenomes[…
03.02.2017 în monitorul oficial nr. Data modificării/datele modificării şi actele normative care modifică (indicând si numărul de ordine a monitoarelor oficiale ce conţin aceste modificări). Jun 28, 2013 (p13) grch38:
Grch38/hg38 is the assembly of the human genome released december of 2013, that uses alternate or alt contigs to represent common complex we strongly recommend switching to grch38/hg38 if you are working with human sequence data. I'd like to provide the gtf to salmon change stdout to the output filename you want in the last command to get an hg19 refgene gtf file: A data set with 24 rows and 5 columns these data derived from the hg38 gap ucsc table, freely available at:

The positions in the files in hg19.zip map to the hg19 reference genome.
Since 2017 june, annovar package now includes hg19_refgenewithver.txt file to give an example how to annotate varians with refgene with versions. 38/2017 din 27 ianuarie 2017. Language english with lithuanian translation.
Grch38/hg38 is the assembly of the human genome released december of 2013, that uses alternate or alt contigs to represent common complex we strongly recommend switching to grch38/hg38 if you are working with human sequence data. Or is there instructions as to how to generate this? I need use ensemble gtf file, hg38.ensgene.gtf.

/u/hg38 has helped pay for 7.46 hours of reddit server time.
03.02.2017 în monitorul oficial nr. Publicat în monitorul oficial nr. 38 din 27 ianuarie 2017pentru aplicarea prevederilor art.

Posting Komentar untuk "Hg 38/2017"